New Paper in Gut Microbes by the Hardt Lab
Mouse models for bacterial enteropathogen infections: insights into the role of colonization resistance
Mathias K-M Herzog, Monica Cazzaniga, Audrey Peters, Nizar Shayya, Luca Beldi, Siegfried Hapfelmeier, Markus M Heimesaat, Stefan Bereswill, Gad Frankel, Cormac G M Gahan, Wolf-Dietrich Hardt
Gut Microbes. 2023 Jan-Dec;15(1):2172667. doi: 10.1080/19490976.2023.2172667.
external page Link to the paper
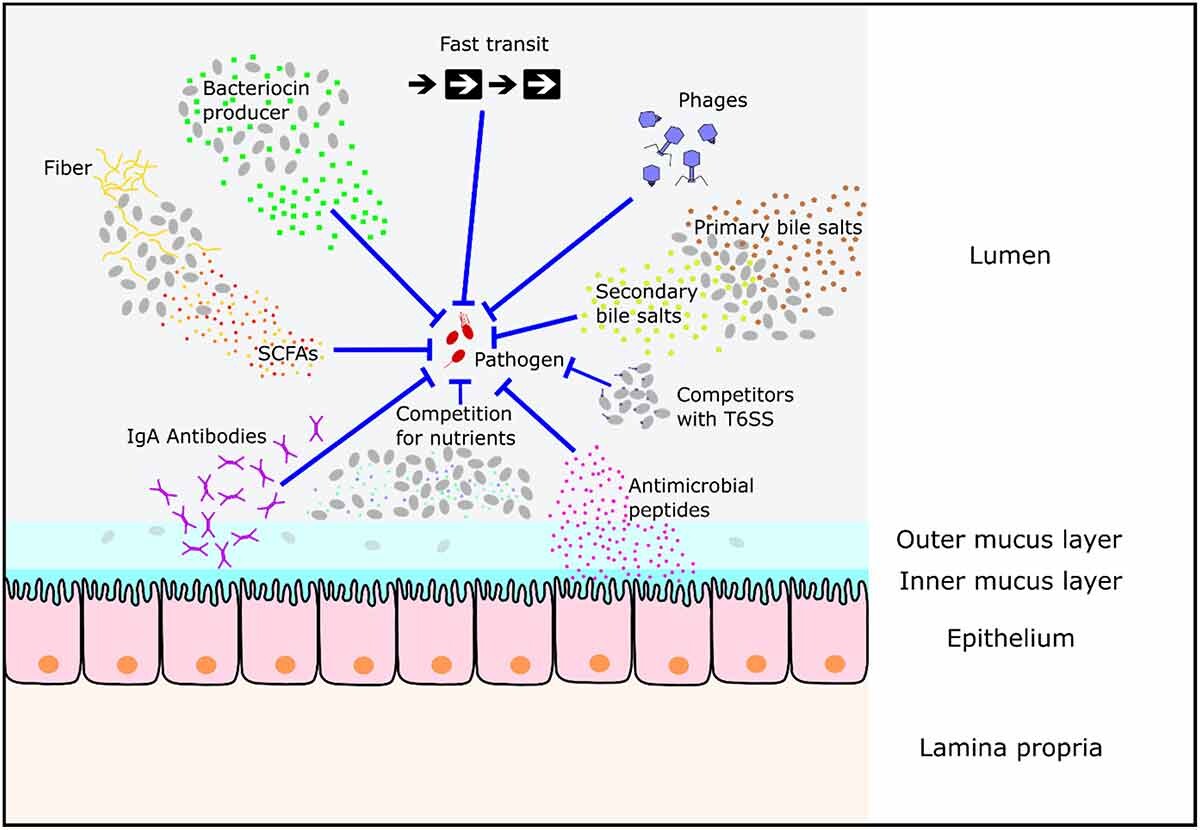
Globally, enteropathogenic bacteria are a major cause of morbidity and mortality.1-3 Campylobacter, Salmonella, Shiga-toxin-producing Escherichia coli, and Listeria are among the top five most commonly reported zoonotic pathogens in the European Union.4 However, not all individuals naturally exposed to enteropathogens go on to develop disease. This protection is attributable to colonization resistance (CR) conferred by the gut microbiota, as well as an array of physical, chemical, and immunological barriers that limit infection. Despite their importance for human health, a detailed understanding of gastrointestinal barriers to infection is lacking, and further research is required to investigate the mechanisms that underpin inter-individual differences in resistance to gastrointestinal infection. Here, we discuss the current mouse models available to study infections by non-typhoidal Salmonella strains, Citrobacter rodentium (as a model for enteropathogenic and enterohemorrhagic E. coli), Listeria monocytogenes, and Campylobacter jejuni. Clostridioides difficile is included as another important cause of enteric disease in which resistance is dependent upon CR. We outline which parameters of human infection are recapitulated in these mouse models, including the impact of CR, disease pathology, disease progression, and mucosal immune response. This will showcase common virulence strategies, highlight mechanistic differences, and help researchers from microbiology, infectiology, microbiome research, and mucosal immunology to select the optimal mouse model.