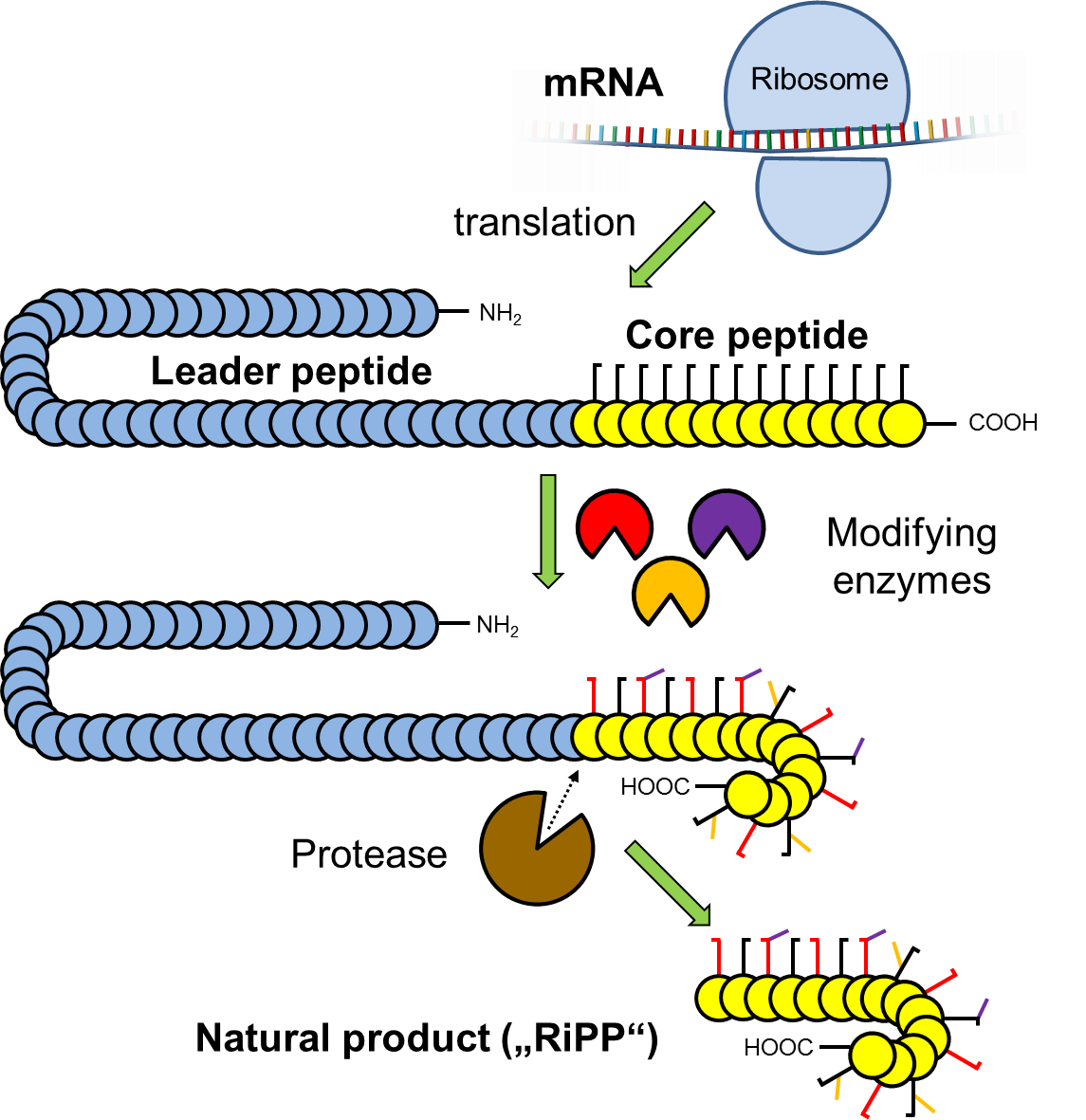
Expanding the biosynthetic scope of peptides
Our studies on uncultivated bacteria revealed RiPP pathways that encode giant peptide toxins of the proteusin-family. Their biosynthesis involves 48 modifications catalyzed by only 6 enzymes. These and further compounds, that we identified by genome mining, belong to various new natural product families that use unprecedented peptide maturases to install for example:
- D-amino acids,
- ornithine,
- fatty acids,
- tert-butyl groups,
- N-methylations,
- and beta-amino acids.
The latter modification was applied in synthetic biology to introduce diverse and multiple beta-amino acids in proteins.
Related publications (selection)
Mordhorst S, Ruijne F, Vagstad AL, Kuipers OP, Piel J. "Emulating nonribosomal peptides with ribosomal biosynthetic strategies." RSC Chem. Biol. (2022) 4, 1, 7-36. external page Abstract
Hubrich F, Lotti A, Scott TA, Piel J. "Uncovering Novel Peptide Chemistry from Bacterial Natural Products." Chimia (Aarau). (2021) 75, 6, 543-547. Abstract
Montalbán-López M, Scott TA, Ramesh S, Rahman IR, van Heel AJ, Viel JH, Bandarian V, Dittmann E, Genilloud O, Goto Y, Grande Burgos MJ, Hill C, Kim S, Koehnke J, Latham JA, Link AJ, Martínez B, Nair SK, Nicolet Y, Rebuffat S, Sahl HG, Sareen D, Schmidt EW, Schmitt L, Severinov K, Süssmuth RD, Truman AW, Wang H, Weng JK, van Wezel GP, Zhang Q, Zhong J, Piel J, Mitchell DA, Kuipers OP, van der Donk WA. "New developments in RiPP discovery, enzymology and engineering." Nat. Prod. Rep. (2021) 38, 1, 130-239. external page Abstract
Scott TA, Piel J. "The hidden enzymology of bacterial natural product biosynthesis." Nat. Rev. Chem. (2019) 7, 404-425. external page Abstract
Freeman MF, Vagstad AL, Piel J. "Polytheonamide biosynthesis showcasing the metabolic potential of sponge-associated uncultivated 'Entotheonella' bacteria." Curr. Opin. Chem. Biol. (2016) 8-14. external page Abstract
Richter D, Lakis E, Piel J. "Site-specific bioorthogonal protein labelling by tetrazine ligation using endogenous β-amino acid dienophiles." Nat. Chem. (2023) doi: 10.1038/s41557-023-01252-8. external page Abstract
Lakis E, Magyari S, Piel J. "In Vivo Production of Diverse β-Amino Acid-Containing Proteins." Angew. Chem. Int. Ed. (2022), 61, 29, e202202695. external page Abstract
Scott TA, Verest M, Farnung J, Forneris CC, Robinson SL, Ji X, Hubrich F, Chepkirui C, Richter DU, Huber S, Rust P, Streiff AB, Zhang Q, Bode JW, Piel J. "Widespread microbial utilization of ribosomal β-amino acid-containing peptides and proteins" Chem (2022) 8, 10, 2659-2677. external page Abstract
Morinaka BI, Lakis E, Verest M, Helf MJ, Scalvenzi T, Vagstad AL, Sims J, Sunagawa S, Gugger M, Piel J. "Natural noncanonical protein splicing yields products with diverse β-amino acid residues." Science. (2018) 359, 6377, 779-782. external page Abstract
Mordhorst S, Badmann T, Bösch NM, Morinaka BI, Rauch H, Piel J, Groll M, Vagstad AL. "Structural and Biochemical Insights into Post-Translational Arginine-to-Ornithine Peptide Modifications by an Atypical Arginase." ACS Chem. Biol. (2023) 18, 3, 528-536. external page Abstract
Hubrich F, Bösch NM, Chepkirui C, Morinaka BI, Rust M, Gugger M, Robinson SL, Vagstad AL, Piel J. "Ribosomally derived lipopeptides containing distinct fatty acyl moieties." Proc. Natl. Acad. Sci. U. S. A. (2022) 119, 3, e2113120119. external page Abstract
Mordhorst S, Morinaka BI, Vagstad AL, Piel J. "Posttranslationally Acting Arginases Provide a Ribosomal Route to Non-proteinogenic Ornithine Residues in Diverse Peptide Sequences." Angew. Chem. Int. Ed. (2020) 59, 48, 21442-21447. external page Abstract
Bösch NM, Borsa M, Greczmiel U, Morinaka BI, Gugger M, Oxenius A, Vagstad AL, Piel J. "Landornamides: Antiviral Ornithine-Containing Ribosomal Peptides Discovered through Genome Mining." Angew. Chem. Int. Ed. (2020) 59, 29, 11763-11768. external page Abstract
Korneli M, Fuchs SW, Felder K, Ernst C, Zinsli LV, Piel J. "Promiscuous Installation of D-Amino Acids in Gene-Encoded Peptides." ACS Synth. Biol. (2021) 10, 2, 236-242. external page Abstract
Vagstad AL, Kuranaga T, Püntener S, Pattabiraman VR, Bode JW, Piel J. "Introduction of D-Amino Acids in Minimalistic Peptide Substrates by an S-Adenosyl-l-Methionine Radical Epimerase." Angew. Chem. Int. Ed. (2019) 58, 8, 2246-2250. external page Abstract
Morinaka BI, Vagstad AL, Piel J. "Radical S-Adenosylmethionine Peptide Epimerases: Detection of Activity and Characterization of D-Amino Acid Products." Methods Enzymol. (2018) 604, 237-257. external page Abstract
Morinaka BI, Verest M, Freeman MF, Gugger M, Piel J. "An Orthogonal D2O-Based Induction System that Provides Insights into D-Amino Acid Pattern Formation by Radical S-Adenosylmethionine Peptide Epimerases." Angew. Chem. Int. Ed. (2017) 56, 3, 762-766. external page Abstract
Fuchs SW, Lackner G, Morinaka BI, Morishita Y, Asai T, Riniker S, Piel J. "A Lanthipeptide-like N-Terminal Leader Region Guides Peptide Epimerization by Radical SAM Epimerases: Implications for RiPP Evolution." Angew. Chem. Int. Ed. (2016) 55, 40, 12330-3. external page Abstract
Morinaka BI, Vagstad AL, Helf MJ, Gugger M, Kegler C, Freeman MF, Bode HB, Piel J "Radical S-adenosyl methionine epimerases: regioselective introduction of diverse D-amino acid patterns into peptide natural products" Angew Chem Int Ed Engl. (2014) 53, 8503-7. external page Abstract
Cogan DP, Bhushan A, Reyes R, Zhu L, Piel J, Nair SK. "Structure and mechanism for iterative amide N-methylation in the biosynthesis of channel-forming peptide cytotoxins." Proc. Natl. Acad. Sci. U. S. A. (2022) 119, 13, e2116578119. external page Abstract
Helf MJ, Freeman MF, Piel J. "Investigations into PoyH, a promiscuous protease from polytheonamide biosynthesis." J. Ind. Microbiol. Biotechnol. (2019) 46, 3-4, 551-563. external page Abstract
Helf MJ, Jud A, Piel J. "Enzyme from an Uncultivated Sponge Bacterium Catalyzes S-Methylation in a Ribosomal Peptide." Chembiochem. (2017) 18, 5, 444-450. external page Abstract
Freeman MF, Helf MJ, Bhushan A, Morinaka BI, Piel J. "Seven enzymes create extraordinary molecular complexity in an uncultivated bacterium." Nat. Chem. (2017) 4, 387-395. external page Abstract
Freeman MF, Gurgui C, Helf MJ, Morinaka BI, Uria AR, Oldham NJ, Sahl HG, Matsunaga S, Piel J. "Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides." Science (2012) 338, 6105, 387-90. external page Abstract