The Piel Lab - Bacterial Natural Products
Prof. Jörn Piel

ETH Zürich
Institut für Mikrobiologie
HCI F 431
Vladimir-Prelog-Weg 4
8093 Zürich
phone: +41 44 633 07 55
email:
Natural products are the basis of many drugs. We aim to understand how bacteria make structurally complex natural products and how to discover metabolic novelty in unexplored microbes. We use this knowledge to generate structural diversity not yet encountered in nature and to create sustainable sources of rare bioactive compounds. To achieve these goals, scientists from diverse backgrounds collaborate in our group, including molecular and microbiology, bioinformatics, and analytical and organic chemistry. For more information, please follow the links above.
Open Positions
We are always looking for motivated new members to join our group (application guideline).
Research Snippets
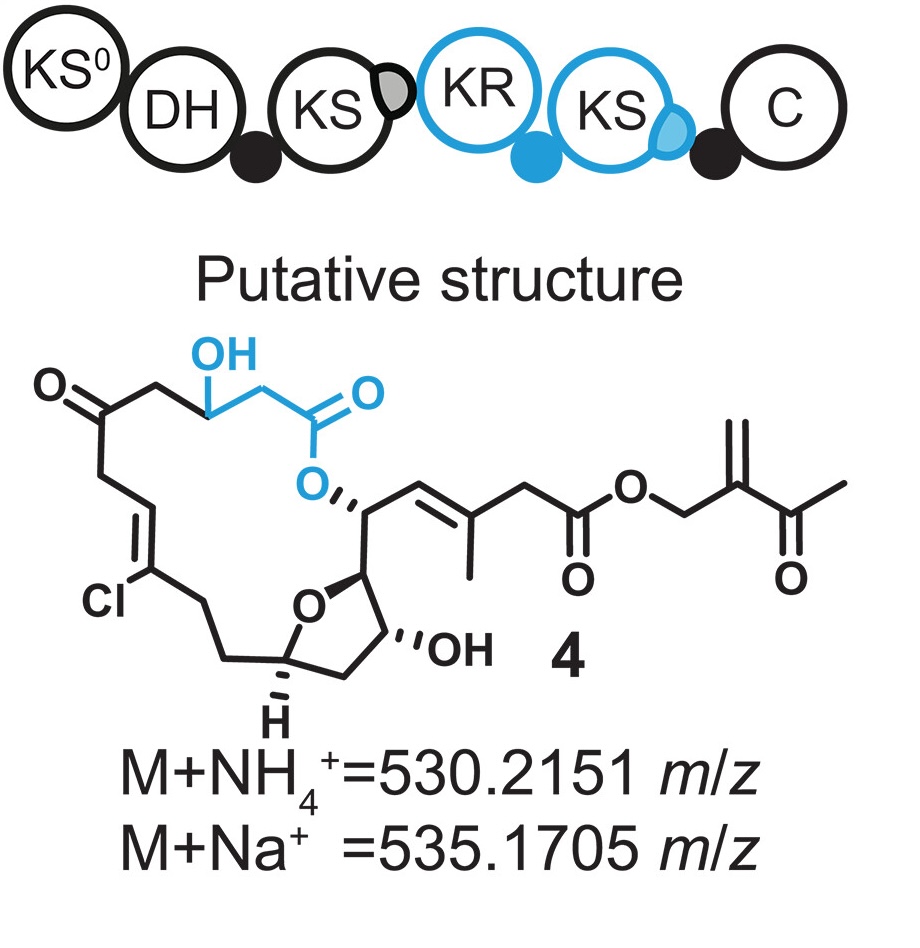
Evolution-guided engineering of trans-acyltransferase polyketide synthases (external page Science)
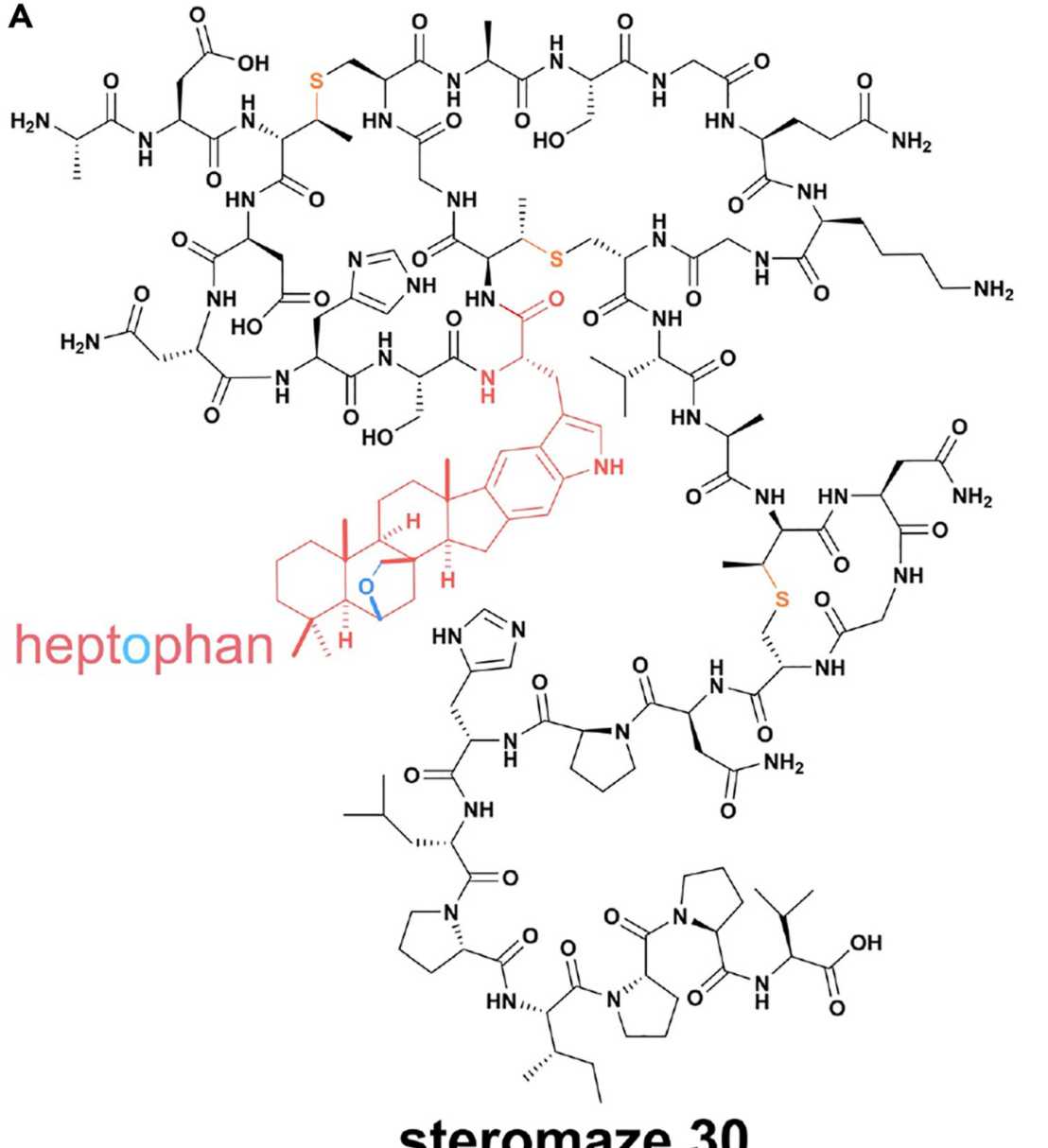
Ribosomal peptides with polycyclic isoprenoid moieties (external page Chem)
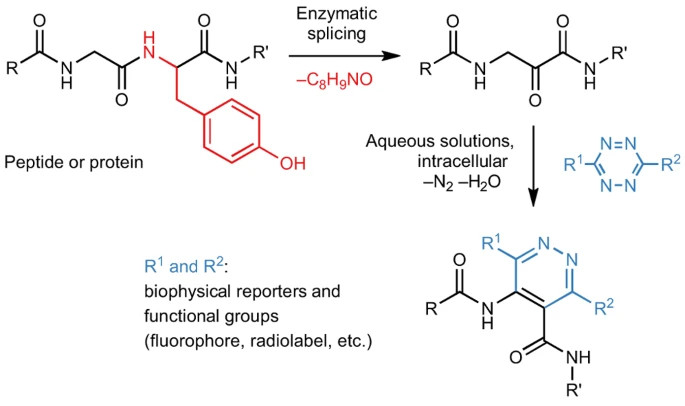
Site-specific bioorthogonal protein labelling by tetrazine ligation using endogenous β-amino acid dienophiles (external page Nature Chemistry)

Animal-associated marine Acidobacteria with a rich natural-product repertoire (external page Chem)external page
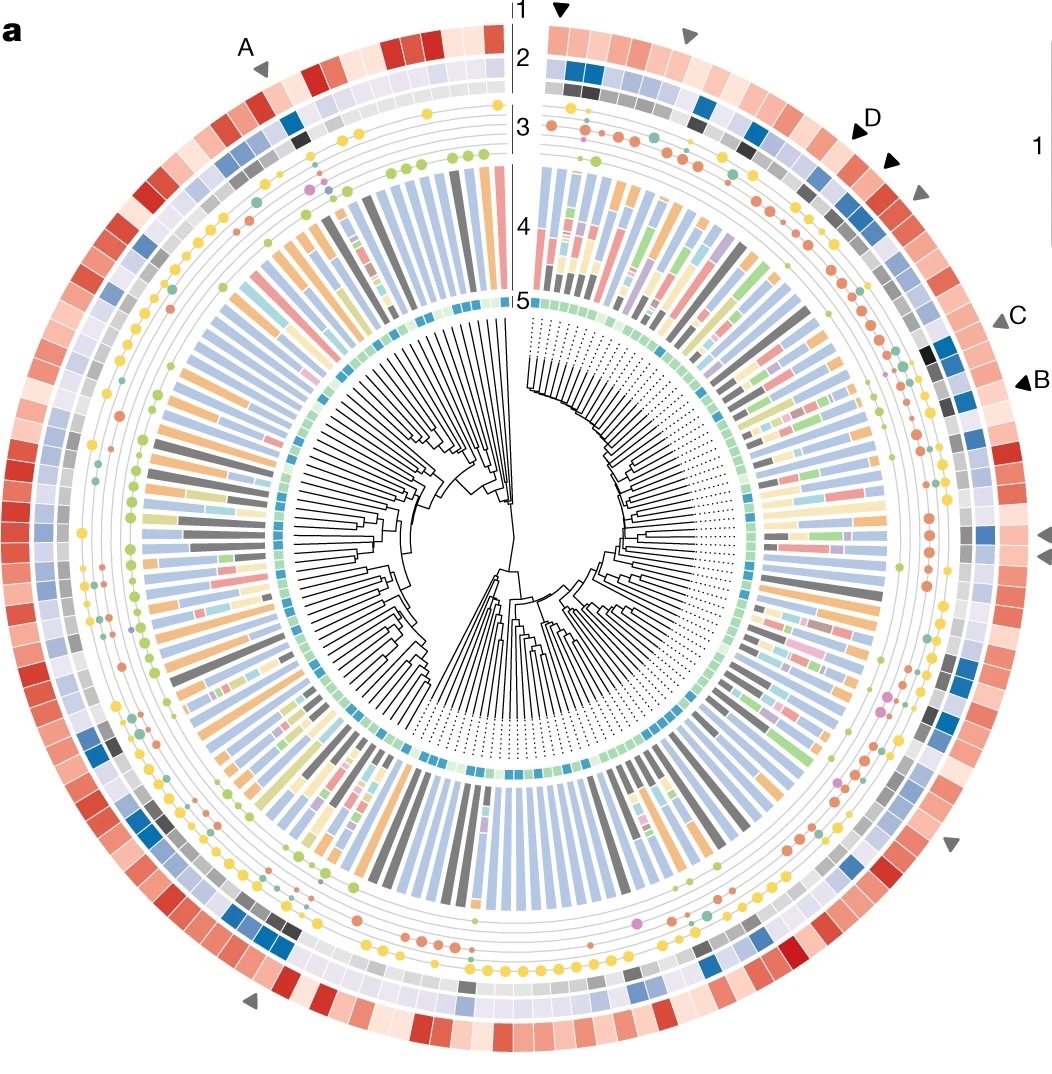
Biosynthetic potential of the global ocean microbiome (external page Nature)
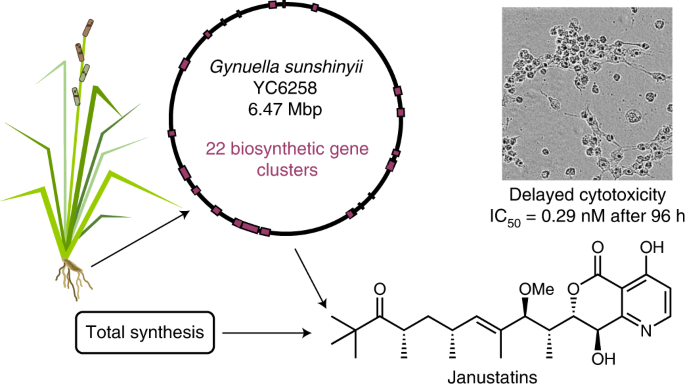
Genome-based discovery and total synthesis of janustatins, potent cytotoxins from a plant-associated bacterium (external page Nature Chemistry)
Alumni Research Groups
Current position: Group leader at Radboud University
external page The Mabesoone lab is leveraging advances in automated synthesis, high-throughput experimentation and machine learning for chemistry to guide the design of biobased soft materials and evolve their properties.
Current position: Junior Professor at Universität Tübingen
external page The Mordhorst lab focuses on peptides from microbial specialised metabolism. We use genome mining to identify novel RiPP biosynthetic gene clusters and investigate these biosynthetic pathways focussing on the biochemical characterisation of the involved enzymes and on their potential for peptide engineering.
Current position: Group Leader (tenure-track) at EAWAG
The external page Microbial Specialized Metabolism group led by Serina Robinson focuses on using multi-omic datasets to gain functional insights into microbial biotransformations. We're interested in combining metagenome mining and machine learning with experimental approaches to characterize enzymes and pathways for pollutant biodegradation and secondary metabolite biosynthesis.
Current position: Assistant Professor at the University of Tokyo
external page Aim of the Synthetic Biology Laboratory: Microorganisms are too small for us to observe with the naked eye. So, we are usually not aware of their presence. But they are in fact all around us. For example, there are many microorganisms under the mulch in the forest, etc., and they also coexist in our bodies. In our laboratory, by discovering infinitely unknown functions of microbial genes and elucidating their basic principles, we aim to create new useful compounds and understand complex interactions between microorganisms.
Current position: Professor at Goethe-Universität and LOEWE-Zentrum
In the external page Helfrich Lab, we develop tools and workflows for the identification of novel bioactive metabolites from a wide variety of bacterial phyla and diverse ecological niches. In addition, we are interested in deciphering the genetic blueprints that govern their biosynthesis and aim at understanding the role of the identified natural products in their native environment.
Current position: Assistant Professor at National University of Singapore
The overall goal of the external page Morinaka Lab is to identify, characterize, and utilize new posttranslational modifying enzymes from natural product pathways for the production of peptide-based therapeutics and biomaterials. Our lab uses a combination of bioinformatics and synthetic biology to identify pathways of interest and to create modified peptides, respectively.
Current position: Assistant Professor at University of Minnesota
The external page Freeman lab is interested in how small molecules known as natural products are made in the environment. We are especially interested in natural products produced by uncultivated microbial ‘dark matter’ and those produced by unconventional bacterial and fungal sources.
Current position: Group Leader at Universität Bonn
The external page Crüsemann group is interested in the discovery and biosynthesis of structurally novel and bioactive natural products from underexplored sources. These insights will help us to engineer these biosynthetic pathways for the generation of novel compounds with improved structural or biological activities.
Current position: Group Leader at Hans-Knöll Institut
At the external page Synthetic Microbiology Research Unit, our aim is the discovery of novel biosynthetic routes useful for pharmacology, biotechnology, or agriculture. Currently, we are investigating new (and rare) cofactors from bacteria, e.g., of mycobacteria. Our goal is therefore to elucidate the biosynthesis and physiological function of the cofactors and to foster their biotechnological use.
Current position: Assistant Professor at external page Hokkaido University
Current position: external page Professor at University of Medicine and Pharmacy Ho Chi Minh City
Current position: external page Professor at Huazhong Agricultural University
Biosynthesis, regulation and resistance mechanism of natural products from microorganisms.
Current position: Professor at the Technical University of Dresden
The external page Gulder group is interested in the (bio-)chemistry of natural products. We are working on the targeted discovery of new natural product structures by applying bioinformatic and synthetic biology techniques, the elucidation of natural product biosynthetic pathways and the biocatalytic application of biosynthetic enzymes in the total synthesis of complex small molecules and technical applications.
Current position: Assistant Professor at external page Kyoto University
Current position: external page Professor at Shandong University
Isolation of microbial strains from different habitats, isolation of secondary metabolites based on product activity guidance, or direct cloning of gene clusters of active natural products and heterologous expression based on gene cluster sequences, or de novo synthesis of natural product gene clusters and heterologous expression, or screening from compound libraries to mine active substances with new structures, new activities or new targets.
Current position: external page Professor at Huazhong University of Science & Technology
