Tools
These methods enabled the discovery of diverse, potent compounds from uncultivated bacteria and poorly studied bacterial lineages.
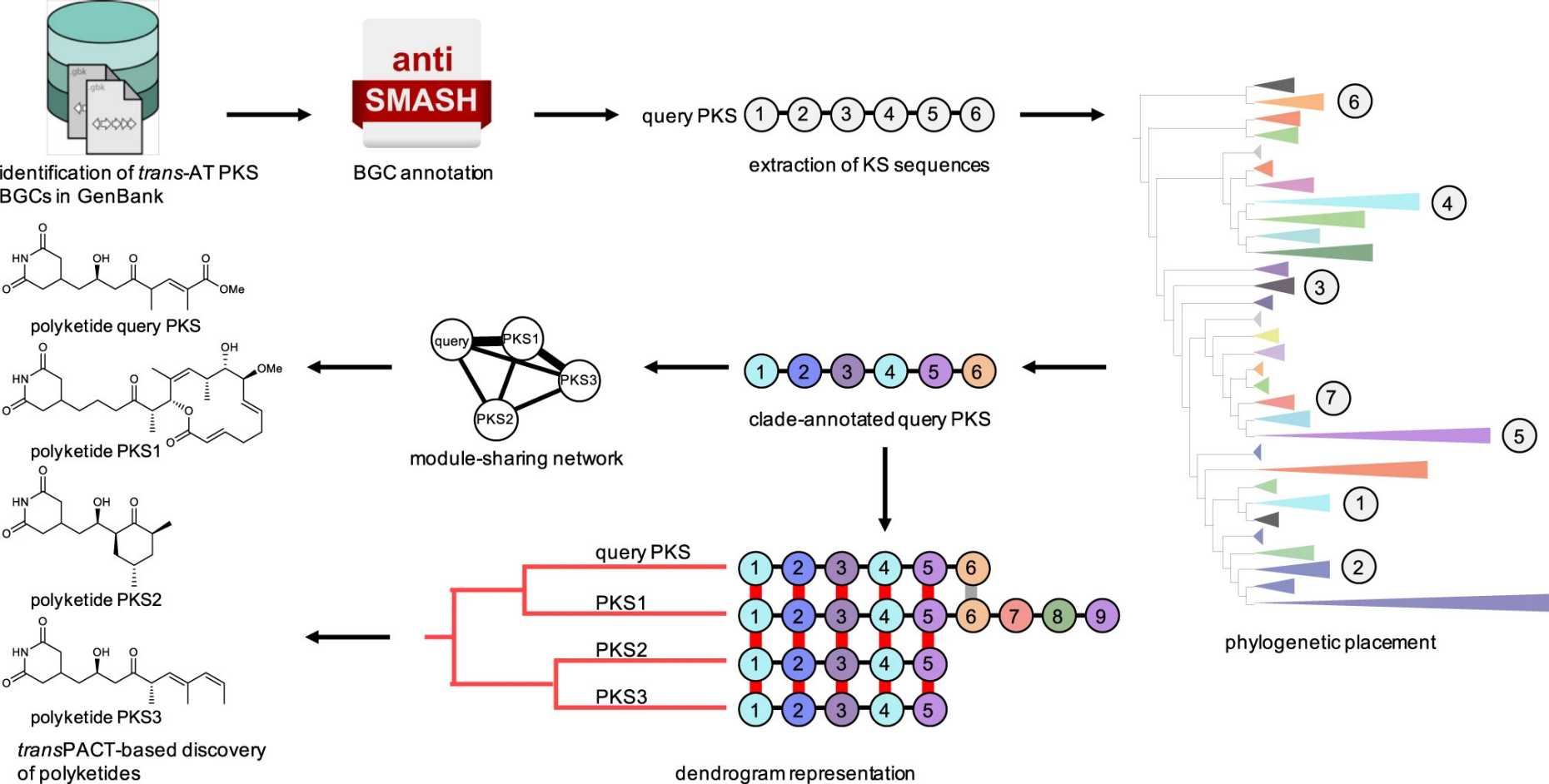
TransPACT
The external page transPACT algorithm (external page Nature Communications) identifies conserved chemical substructures in trans-AT PKS-derived polyketides. We used it to study evolutionary patterns across bacterial genomes and to discover polyketide variants with conserved moieties.
- Code at external page GitHub
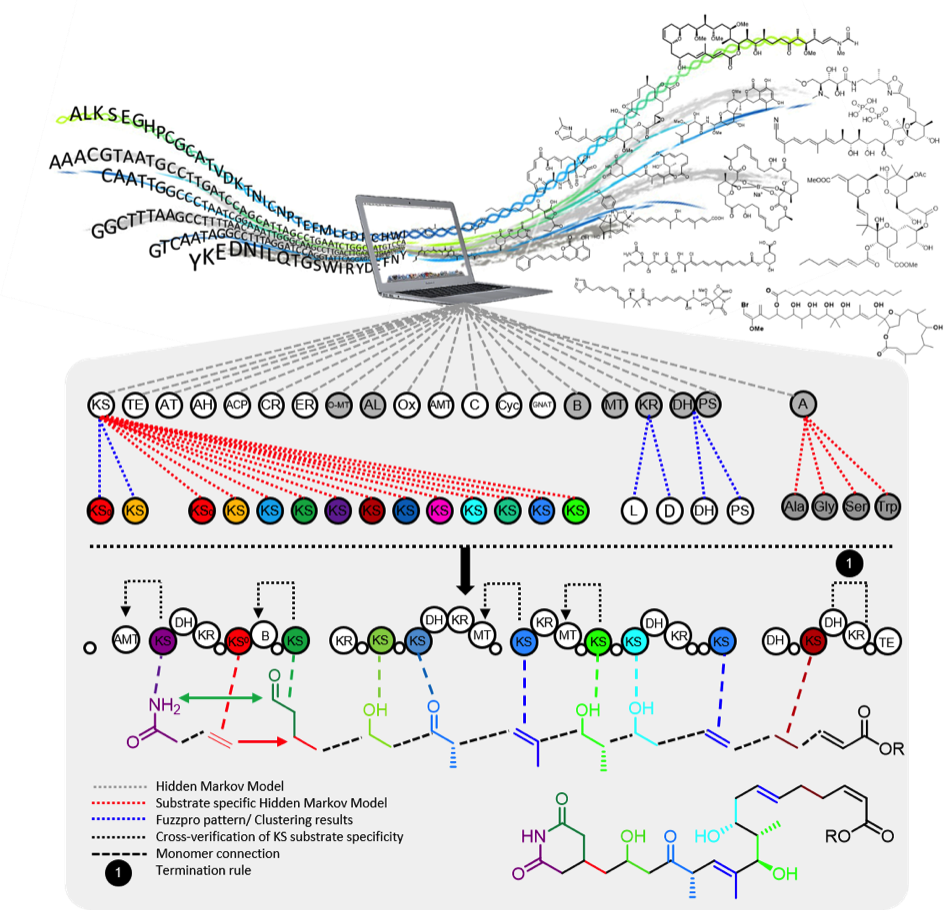
TransATor
The TransATor tool (external page Nature Chemical Biology) makes structure predictions for trans-AT PKS-derived polyketides from protein sequences. We applied it for hypothesis-based polyketide discovery and biosynthetic studies.
- Free TransATor web application at transator.ethz.ch/trans-AT-PKS/
- Code and tutorial at external page GitHub
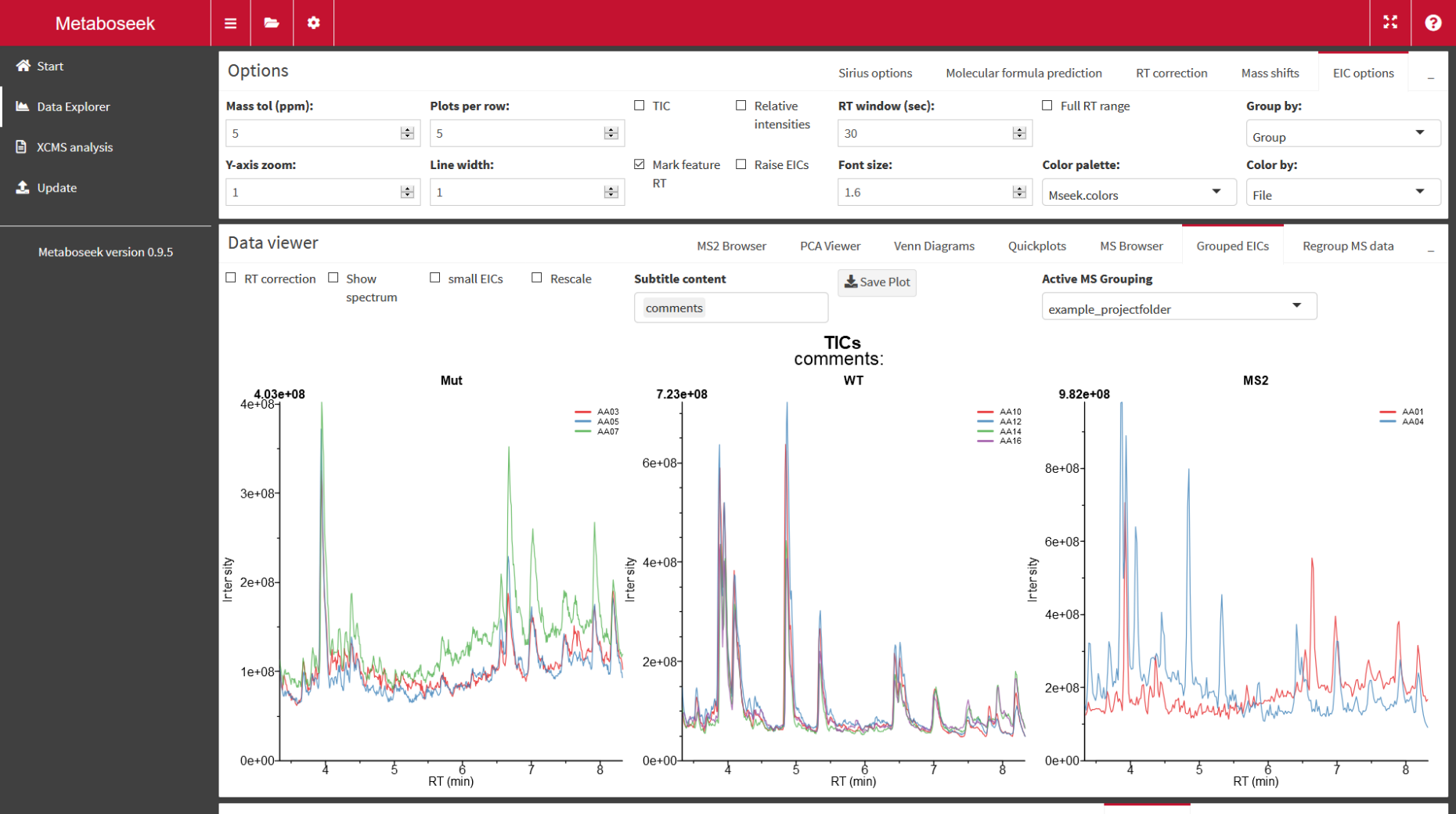
Metaboseek
external page Metaboseek is a tool to analyze LC/MS data. Molecular features are detected and compared across mutliple samples. Metaboseek was developed by former PhD student Dr. Maximilian Helf at Cornell University (external page Nature Communications).
- Free Metaboseek web application at external page metaboseek.com
- Code at external page GitHub