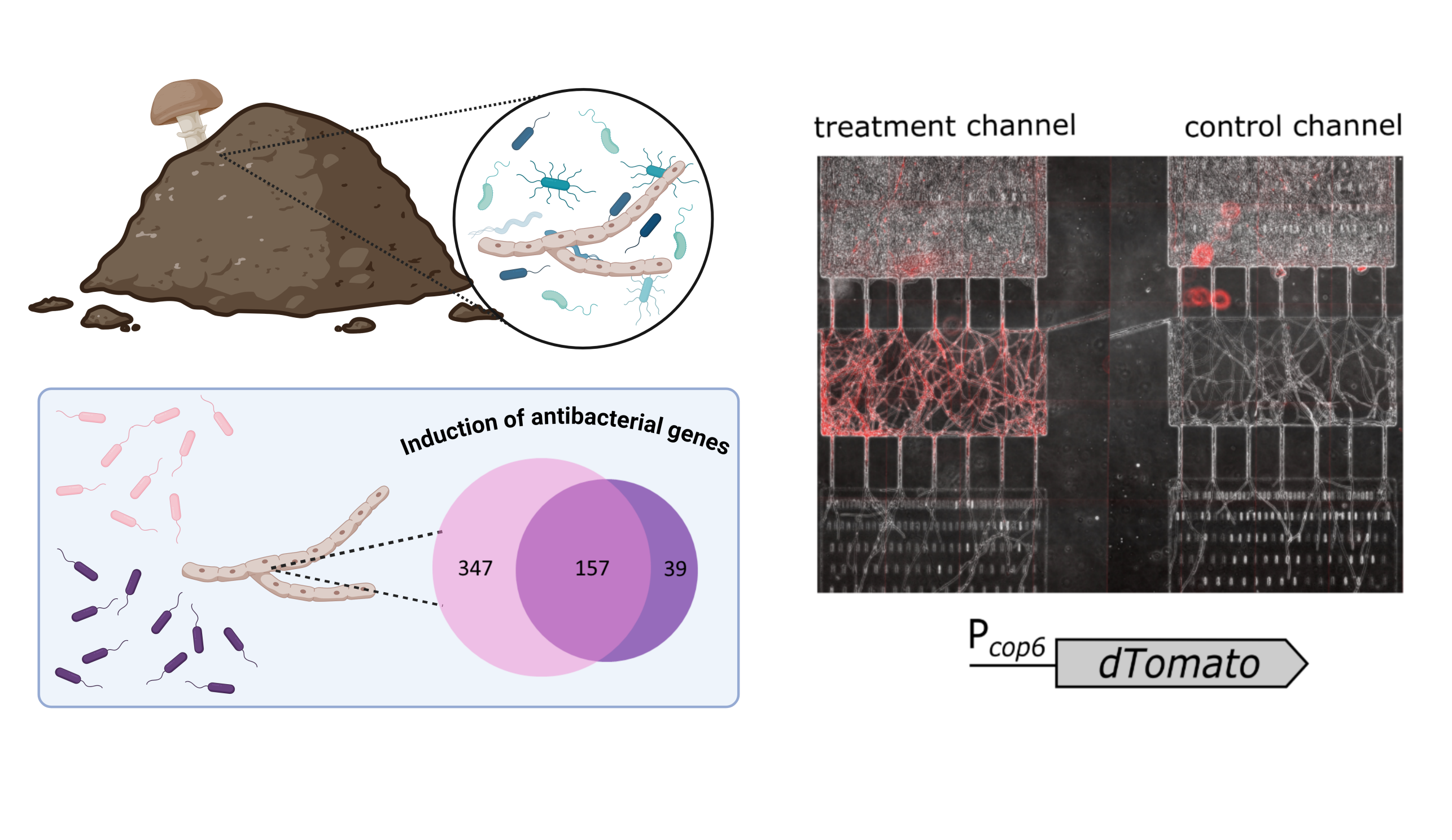
Project 2: Fungal defense against bacteria
Fungi and bacteria are phylogenetically distant, yet they often share ecological niches, which can result in competition over limited resources such as space and nutrients.The constitutive and inducible layers of antibacterial defense are poorly characterized in fungi. In particular, little is known about how fungi sense the presence of antagonistic bacteria in their surroundings and how sensing is eventually translated into a specific defense response.
Our recent findings indicate that antibacterial defence in the model mushroom Coprinopsis cinerea is not only triggered by living bacteria but also upon detection of bacteria-derived soluble compounds. Specifically, the cell-free supernatant from axenic cultures of either Gram-negative Escherichia coli or Gram-positive Bacillus subtilis is sufficient for the induction of genes encoding antibacterial products.
This project aims at:
1) Identifying the bacterial elicitor, or elicitors, of antibacterial defence in C. cinerea.
2) Characterizing the fungal molecular components required for sensing and downstream signalling.
3) Determining whether the antibacterial defence response is conserved across fungal phyla.
To address the above questions, we apply different tools and methodologies ranging from microfluidics devices to RNA-Seq to proteomics. We also develop fungal reporter strains to monitor the induction of antibacterial defense by fluorescence or enzymatic activity.