The Sunagawa Lab - Microbiome Research
Prof. Shinichi Sunagawa

ETH Zürich
Institut für Mikrobiologie
HCI F 417
Vladimir-Prelog-Weg 4
8093 Zürich
How to find us [external page link]
email:
Our lab investigates microbial communities across diverse ecosystems, with a focus on the ocean, aerosols at the land-ocean interface, and the gastrointestinal tract. To support this work, we build genomic data resources, develop taxon-centric tools, and integrate genome-resolved meta’omic data with contextual environmental information. By combining computational approaches with experimental methods, we aim to uncover the ecological and evolutionary drivers of community diversity and function, as well as the mechanisms underlying microbial interactions. We collaborate with interdisciplinary groups, participate in research consortia, and strive to translate insights from basic research into conservation initiatives and public education.
Selected publications

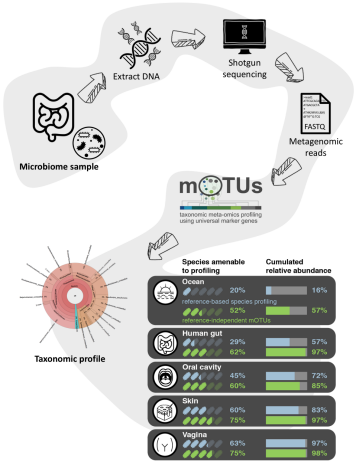
The mOTUs online database provides web-accessible genomic context to taxonomic profiling of microbial communities
Dmitrijeva M*, Ruscheweyh HJ*, Feer L, Li K, Miravet-Verde S, Sintsova A, Mende DR, Zeller G, Sunagawa S✉
external page Nucleic Acids Res: gkae1004, 2024
Cultivation-independent genomes greatly expand taxonomic-profiling capabilities of mOTUs across various environments
Ruscheweyh HJ*, Milanese A*, Paoli L, Karcher N, Clayssen Q, Keller MI, Wirbel J, Bork P, Mende DR, Zeller G✉, Sunagawa S✉
external page Microbiome: 10, 212, 2022

Biosynthetic potential of the global ocean microbiome
Paoli L, Ruscheweyh HJ*, Forneris CC*, Hubrich F*, Kautsar S, Bhushan A, Lotti A, Clayssen Q, Salazar G, Milanese A, Carlstrom CI, Papadopoulou C, Gehrig D, [...], Robinson SL✉, Piel J✉, Sunagawa S✉
external page Nature: 607(7917), 111-118, 2022
Exact genomic locations of inducible prophages and accurate phage-to-host ratios in gut microbial strains
Zünd M, Ruscheweyh HJ, Field CM, Meyer, N, Cuenca M, Hoces D, Hardt WD, Sunagawa S✉
external page Microbiome: 9, 77, 2021

Tara Oceans: towards global ocean ecosystems biology
Sunagawa S✉, Acinas SG, Bork P, Bowler C, Tara Oceans Coordinators, Eveillard D, Gorsky G, Guidi L, Iudicone D, Karsenti E, Lombard F, Ogata H, Pesant S, Sullivan MB, Wincker P, de Vargas C✉
external page Nat Rev Microbiol: 18(8), 428-445, 2020

Gene expression changes and community turnover differentially shape the global ocean metatranscriptome
Salazar G*, Paoli L*, Alberti A, Huerta-Cepas J, Ruscheweyh HJ, Cuenca M, Field CM, Coelho LP, [...] and Sunagawa S✉
external page Cell: 179(5), 1068-1083, 2019

Structure and function of the global ocean microbiome
Sunagawa S*✉, Coelho LP*, Chaffron S*, Kultima JR, Labadie K, Salazar G, [...], Wincker P, Karsenti E, Raes J✉, Acinas SG✉, Bork P✉
external page Science: 348(6237), 1261359, 2015



