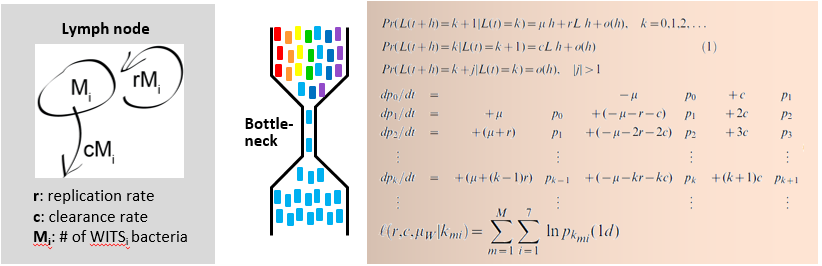
Salmonella population dynamics
In an infection, the pathogen has to successfully complete a series of interactions with the physical/chemical barriers, the resident microbiota, the different layers of the infected tissue and the immune defences of the host. Many of these steps are performed only by a small subpopulation of the pathogen, thus creating "colonization bottlenecks" (Kaiser et al., 2013; Maier et al., 2014). The mechanisms limiting the critical steps of an infection are of great interest, as they represent "Achilles's heels" of value for cure or prevention. In collaboration with Roland Regoes, we are combining barcoded strains, mouse infection assays and population dynamics models to identify bottlenecks and decipher their molecular or cellular nature.
Recently, we have employed this approach to assess why standard antibiotic therapies fail to cure Salmonella diarrhea. We found that bacterial persistence represents a key. The persistent bacteria are formed in vivo as a result of the invasion into particular types of host cells, i.e. dendritic cells of the gut associated immune system. Within these cells, S. Typhimurium forms a subpopulation of slow-growing antibiotic tolerant "persister" cells (Kaiser et al., 2014; Diard et al., 2014). These persisters survive an antibiotic therapy and cause relapses once the therapy is discontinued. Our system should provide an excellent starting point for studying the pathogen-host cell interactions driving persister formation in vivo and for developing strategies to eliminate persisters.
The barcoding approach can also be applied to estimate transfer-rates of resistance plasmids and phages between different bacterial strains in the gut. This technique revealed that the inflamed intestine fosters extremely high rates of horizontal gene transfer between Enterobacteriaceae (Diard et al., Science 2017).
Literature
Kaiser, P., Slack, E., Grant, A.J., Hardt, W.D. and R.R. Regoes (2013) Lymph node colonization dynamics after oral Salmonella Typhimurium infection in mice, PLoS Pathogens, 9(9):1-12. external page [Abstract]
Kaiser, P., Regoes, R.R., Dolowschiak, T., Wotzka, S., Lengefeld, J., Slack, E., Grant, A.J., Ackermann, M. and W.D. Hardt (2014) Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment, PLoS Biology, 12(2):e1001793. external page [Abstract]
Maier, L., Diard, M., Sellin, M.E., Chouffane, E.S., Trautwein-Weidner, K., Periaswamy, B., Slack, E., Dolowschiak, T., Stecher, B., Loverdo, C., Regoes, R.R., and W.D. Hardt* (2014) Granulocytes Impose a Tight Bottleneck upon the Gut Luminal Pathogen Population during Salmonella Typhimurium Colitis, PLoS Pathogens, 10(12), e1004557. external page [Abstract]
Kaiser, P., Regoes R.R. and W.D. Hardt (2016) Population Dynamics Analysis of Ciprofloxacin-Persistent S. Typhimurium Cells in a Mouse Model for Salmonella Diarrhea, Methods Mol Biol., 1333:189-203. external page [Abstract]
Diard, M., Bakkeren, E., Cornuault, J. K, Moor, K., Hausmann, A., Sellin, M. E., Loverdo, C., Aertsen, A., Ackermann, M., De Paepe, M., Slack, E., Hardt, W.D., (2017) Inflammation boosts bacteriophage transfer between Salmonella spp., Science, 6330:1211-1215. external page [Abstract]